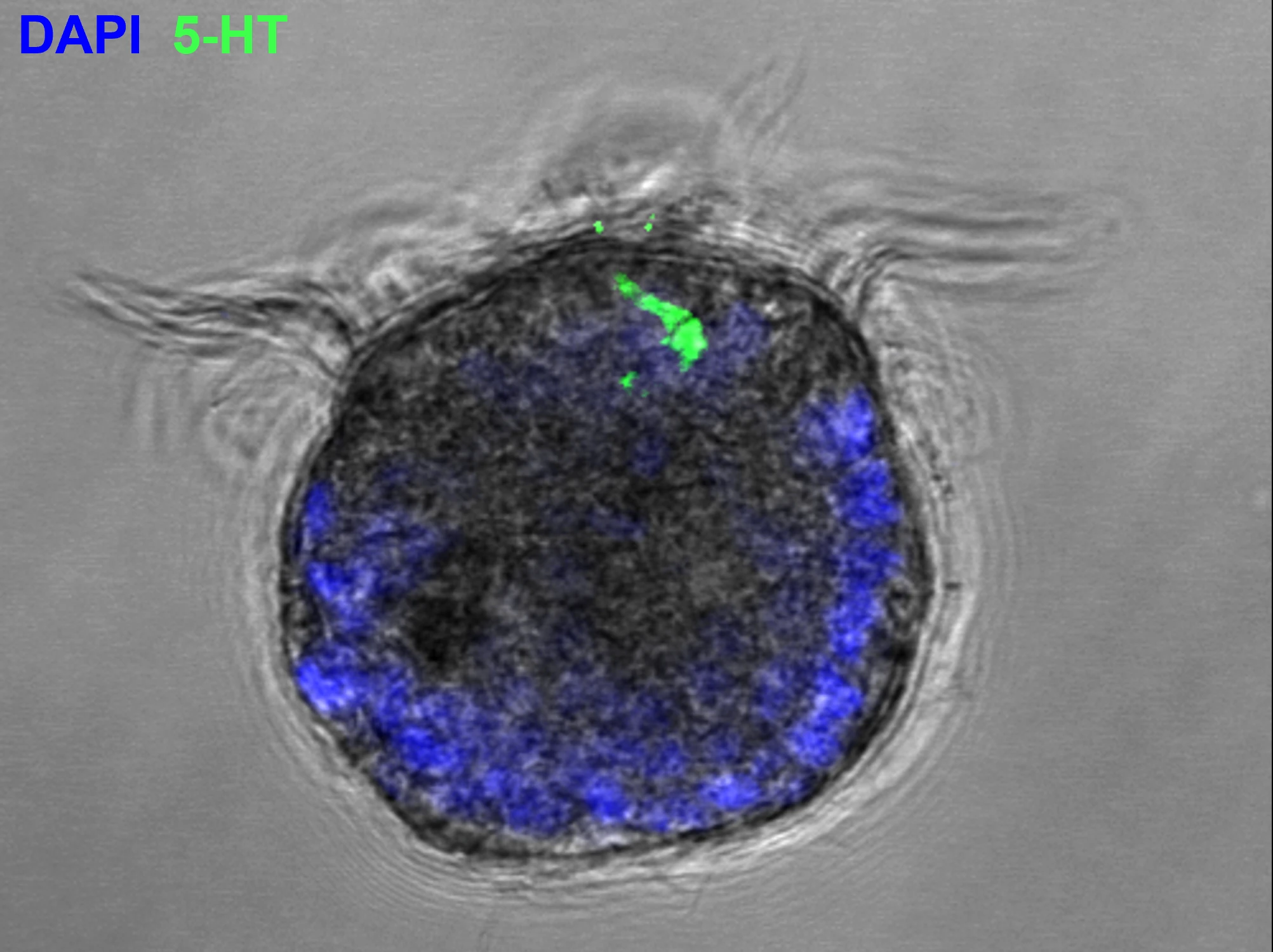
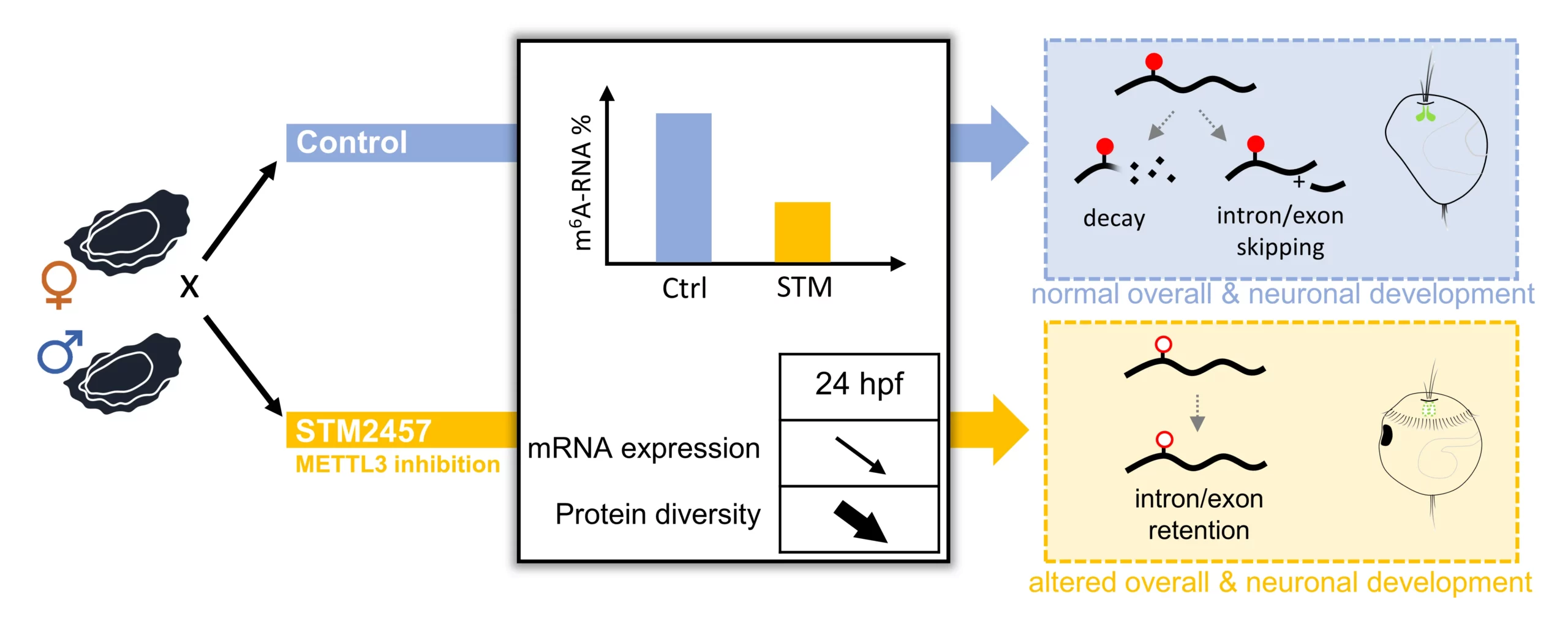
m6A-RNA epitranscriptomes regulate splicing and neuronal development in the Pacific oyster Crassostrea gigas
Natacha Clairet, Hélène Auger, Roberto-Carlos Arredondo-Espinoza, Hugo Koechlin, Benoît Bernay, Lukas Manoury, Didier Goux, Guillaume Rivière.
N6-adenosine RNA methylation (m6A) is a key regulator of gene expression during embryogenesis and neurogenesis in mammals and insects. However, its functional relevance remains unknown in lophotrochozoans like the Pacific oyster Crassostrea gigas, despite its association with developmental gene expression.
We treated oyster embryos with STM2457, a METTL3 methyltransferase-inhibitor. m6A-RNA reduction in treated embryos induced morphological alterations and 5-HT immunohistochemistry revealed impaired neuronal development. Transcriptome and proteome analyses indicated that m6A inhibition disturbs transcription and translation. Epitranscriptome sequencing showed that m6A inhibition increased transcript length by exon and intron retention, suggesting m6A-dependent recruitment of splicing factors at intron–exon boundaries.
Together, our results support an essential role for m6A in neural differentiation and development in the oyster by regulating alternative splicing. This study provides the first evidence of a functional role for m6A in lophotrochozoan development, providing new eco-evo-devo insights of epitranscriptomic processes.